DIGLM Tutorial¶
In this notebook we illustrate with a toy model the usage of the DIGLM class.
The notebook is organized as follows:
Introduction;
Creation of a toy dataset;
Building the model and defining training functons;
Support function for plotting and sampling;
Training;
Results and final considerations
1. Introduction¶
An object of the DIGLM class is a trainable model which allows to accomplish the two following tasks:
Reproduce the feature distribution thanks to a normalizing flow step mapping the features to a latent space with multivariate normal distribution features;
Perform regression (linear, logistic and much more) with a Generalized Linear Model (GLM).
The idea comes from the article by E. Nalisnick et al. where the DIGLM model and algorithm characteristics is described and test on some examples are provided.
The two parts that constitute the algorithm are trained in a single feed-forward step of a semi-supervised training: given the features observation $ {x}_i ^N $ and their true labels $ {y}_i ^N $, the loss function minimized is
where:
\(\beta\) are the weights of the glm;
\(\theta\) are the parameters that maps the distibution of \(x\) to the space of the latent variables \(z\) through the normalizing flow. The distribution of \(z\) is set to be a multivariate normal distribution with dimension equal to the space of the feature vectors;
\(\lambda\) is a hyper-parameter which weights the importance in the training of the two different parts of the loss: the first term indeed is used to solve the regression problem; the second one trains the normalizing flow.
[2]:
import sys
sys.path.append('../../../src')
import os
import logging
import tensorflow as tf
from tensorflow import Variable, ones, zeros
from tensorflow_probability import glm
from tensorflow_probability import distributions as tfd
from tensorflow.keras import metrics
import numpy as np
import seaborn as sns
import sklearn.datasets as skd # where we can find a simple example
import matplotlib.pyplot as plt
import pandas as pd
from diglm import Diglm
from spqr import NeuralSplineFlow as NSF
# sinlencing tensorflow warnings
os.environ['TF_CPP_MIN_LOG_LEVEL'] = '3'
logging.getLogger('tensorflow').setLevel(logging.FATAL)
Cloning into 'diglm'...
remote: Enumerating objects: 828, done.
remote: Counting objects: 100% (220/220), done.
remote: Compressing objects: 100% (76/76), done.
remote: Total 828 (delta 169), reused 162 (delta 144), pack-reused 608
Receiving objects: 100% (828/828), 10.93 MiB | 3.40 MiB/s, done.
Resolving deltas: 100% (476/476), done.
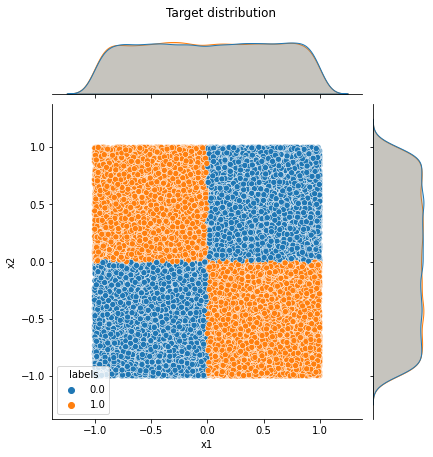
2. Toy dataset¶
The feature vectors \(x_1\) and \(x_2\) are sampled from a uniform distribution \(U[-1, 1]\). The response function \(y(x_1, x_2): \mathbb{R}^2 \rightarrow \{0, 1\}\) is: $
$
The sampled vectors and the response are transformed to a tf.data.Dataset.
[8]:
#response function
response = lambda x1, x2: x1 * x2 > 1 / (x1 * x2)
BATCH_SIZE = 1024
DATASET_SIZE = BATCH_SIZE * 32
VAL_SIZE = BATCH_SIZE * 16
# dataset for training
X1 = np.random.uniform(low=-1, high=1, size=DATASET_SIZE)
X2 = np.random.uniform(low=-1, high=1, size=DATASET_SIZE)
df = pd.DataFrame(np.stack((response(X1, X2), X1, X2), axis=1),
columns=['labels','x1', 'x2'])
# dataset for validation
X1_val = np.random.uniform(low=-1, high=1, size=VAL_SIZE)
X2_val = np.random.uniform(low=-1, high=1, size=VAL_SIZE)
df_val = pd.DataFrame(np.stack((response(X1_val, X2_val), X1_val, X2_val), axis=1),
columns=['labels','x1', 'x2'])
# dataset for test
X1_test = np.random.uniform(low=-1, high=1, size=VAL_SIZE)
X2_test = np.random.uniform(low=-1, high=1, size=VAL_SIZE)
df_test = pd.DataFrame(np.stack((response(X1_test, X2_test), X1_test, X2_test), axis=1),
columns=['labels','x1', 'x2'])
# plotting the toy dataset with marginalized feature distribution too
joint_plot = sns.jointplot(data=df, x='x1', y='x2', hue='labels')
joint_plot.figure.suptitle('Target distribution', y = 1.05)
[8]:
Text(0.5, 1.05, 'Target distribution')
3. Utilities: transforming datasets, sampling of distributions and plotting¶
We define some functions to transform our data into TesnorFlow datasets for automatic batching and parallelization; to extract sample from the transformed distributions and to plot them.
[9]:
def to_tf_dataset(df, columns, dtype='float32', batch_size=32):
"""
util function to transform a pandas.DataFrame into a
batched tf.data.Dataset.
"""
tf_ds = tf.data.Dataset.from_tensor_slices(df[columns].astype(dtype))
tf_ds = tf_ds.prefetch(tf.data.experimental.AUTOTUNE)
tf_ds = tf_ds.cache()
tf_ds = tf_ds.batch(batch_size)
return tf_ds
def sampling(size=500):
"""
returns a sample of the transformed disribution and its computed
response through glm evaluation.
"""
trans_sample = diglm.sample(size)['features']
mean_sample, var_sample, grad_sample = diglm(trans_sample)
x1, x2 = np.split(trans_sample.numpy(), 2, axis=1)
x1 = np.concatenate(x1)
x2 = np.concatenate(x2)
mean = np.concatenate(mean_sample.numpy().T)
return mean, x1, x2
def plot(name, counter=None):
"""
plotting function that gets some samples from the norm. flow
inverse distribution and the calculated response and plots it
alongside with the target distribution
"""
# Get sampling data to plot
mean, x1, x2 = sampling(size=VAL_SIZE)
df_train = pd.DataFrame(np.stack((mean, x1, x2), axis=1),
columns=['labels', 'x1', 'x2'])
#plot
ax_1 = sns.scatterplot(data=df_train, x='x1', y='x2', hue='labels', palette='coolwarm')
ax_1.set_ylim(min(x1), max(x1))
ax_1.set_xlim(min(x1), max(x2))
if counter is not None: # used to track the training epoch
ax_1.set_title(f'Training epoch = {counter}')
plt.savefig(name)
train_ft = to_tf_dataset(df, ['x1', 'x2'], batch_size=BATCH_SIZE)
train_l = to_tf_dataset(df, ['labels'], dtype='int32', batch_size=BATCH_SIZE)
val_ft = to_tf_dataset(df_val, ['x1', 'x2'], batch_size=VAL_SIZE)
val_l = to_tf_dataset(df_val, ['labels'], dtype='int32', batch_size=VAL_SIZE)
test_ft = to_tf_dataset(df_test, ['x1', 'x2'], batch_size=VAL_SIZE)
test_l = to_tf_dataset(df_test, ['labels'], dtype='int32', batch_size=VAL_SIZE)
4. Building the DIGLM¶
We initialize our diglm model with:
a NeuralSplineFlow object as bijector with two splits (there are only two variables
a
tf.glm.Bernoulli()as glm to realize the logistic regression.
[10]:
my_glm = glm.Bernoulli()
neural_spline_flow = NSF(splits=2, spline_params=dict())
diglm = Diglm(neural_spline_flow, my_glm, 2)
train_dict = [{'labels': batch_l,
'features': batch_ft}
for batch_l, batch_ft in zip(train_l, train_ft)]
val_dict = [{'labels': batch_l,
'features': batch_ft}
for batch_l, batch_ft in zip(val_l, val_ft)]
test_dict = [{'labels': batch_l,
'features': batch_ft}
for batch_l, batch_ft in zip(test_l, test_ft)]
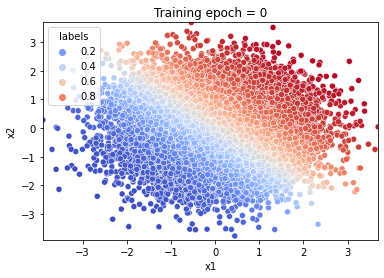
# let's visualize a sample from the inverse distribution of the bijector
# before training.
plot('start.jpeg', counter=0)
5. Training¶
First, we define a train step, where the loss and its gradient are calculated and the training variables of the model are updated accordingly. We follow the keras tutorial.
[11]:
@tf.function
def train_step(optimizer, target_sample, weight=.1):
"""
Train step function for the diglm model. Implements the basic steps for computing
and updating the trainable variables of the model. It also
calculates the loss on training and validation samples.
"""
with tf.GradientTape() as tape:
# calculating loss and its gradient of training data
loss = -tf.reduce_mean(diglm.weighted_log_prob(target_sample, scaling_const=weight))
variables = tape.watched_variables()
gradients = tape.gradient(loss, variables)
optimizer.apply_gradients(zip(gradients, variables))
return loss
Now we are ready to train the model!
[12]:
# we define a suitable metric
accuracy = metrics.BinaryAccuracy()
def update_accuracy():
"""
util function to update the binary accuracy metric
to evaluate the algorithm performances.
"""
accuracy.reset_state()
y_true = test_dict[0]['labels']
y_pred, y_var, grad = diglm(test_dict[0]['features'])
accuracy.update_state(y_true, y_pred.numpy())
history['accuracy'].append(accuracy.result().numpy())
# hyper parameters
LR = 5e-3
NUM_EPOCHS = 500
learning_rate = tf.Variable(LR, trainable=False)
optimizer = tf.keras.optimizers.Adam(learning_rate)
loss = 0
val_loss = 0
history = {'accuracy': [], 'train_loss': [], 'val_loss': []}
fig_list = []
weight = 1
update_accuracy()
print(accuracy.result().numpy())
# training loop on epochs
for epoch in range(NUM_EPOCHS):
if epoch == int(NUM_EPOCHS / 2):
weight = 0.5
if epoch % 10 == 0:
# plot
name = f'fig{epoch}.jpeg'
fig_list.append(name)
plot(name, counter=(epoch + 1))
plt.close()
print(f'Epoch = {epoch+1} \t Loss = {loss:.3f} \t Val loss = {val_loss:.3f}')
# train batch per batch
for train_batch, val_batch in zip(train_dict, val_dict):
loss = train_step(optimizer, train_batch, weight=weight)
# validation loss
val_loss = -tf.reduce_mean(diglm.weighted_log_prob(val_batch, scaling_const=weight))
# updating loss and accuracy
update_accuracy()
history['train_loss'].append(loss)
history['val_loss'].append(val_loss)
0.49371338
Epoch = 1 Loss = 0.000 Val loss = 0.000
Epoch = 11 Loss = 2.908 Val loss = 2.868
Epoch = 21 Loss = 2.808 Val loss = 2.910
Epoch = 31 Loss = 2.614 Val loss = 2.814
Epoch = 41 Loss = 2.413 Val loss = 2.779
Epoch = 51 Loss = 2.422 Val loss = 2.807
Epoch = 61 Loss = 2.324 Val loss = 2.676
Epoch = 71 Loss = 2.400 Val loss = 2.614
Epoch = 81 Loss = 2.252 Val loss = 2.629
Epoch = 91 Loss = 2.161 Val loss = 2.480
Epoch = 101 Loss = 2.108 Val loss = 2.503
Epoch = 111 Loss = 2.052 Val loss = 2.484
Epoch = 121 Loss = 2.043 Val loss = 2.426
Epoch = 131 Loss = 2.106 Val loss = 2.431
Epoch = 141 Loss = 2.046 Val loss = 2.455
Epoch = 151 Loss = 1.982 Val loss = 2.369
Epoch = 161 Loss = 1.975 Val loss = 2.333
Epoch = 171 Loss = 1.969 Val loss = 2.400
Epoch = 181 Loss = 1.915 Val loss = 2.365
Epoch = 191 Loss = 1.904 Val loss = 2.309
Epoch = 201 Loss = 1.863 Val loss = 2.310
Epoch = 211 Loss = 1.842 Val loss = 2.255
Epoch = 221 Loss = 1.884 Val loss = 2.220
Epoch = 231 Loss = 1.847 Val loss = 2.205
Epoch = 241 Loss = 1.908 Val loss = 2.248
Epoch = 251 Loss = 1.821 Val loss = 2.231
Epoch = 261 Loss = 1.087 Val loss = 1.300
Epoch = 271 Loss = 1.042 Val loss = 1.261
Epoch = 281 Loss = 1.031 Val loss = 1.249
Epoch = 291 Loss = 1.072 Val loss = 1.222
Epoch = 301 Loss = 1.048 Val loss = 1.244
Epoch = 311 Loss = 1.012 Val loss = 1.217
Epoch = 321 Loss = 0.976 Val loss = 1.199
Epoch = 331 Loss = 1.046 Val loss = 1.176
Epoch = 341 Loss = 0.987 Val loss = 1.207
Epoch = 351 Loss = 1.004 Val loss = 1.149
Epoch = 361 Loss = 0.981 Val loss = 1.146
Epoch = 371 Loss = 0.969 Val loss = 1.130
Epoch = 381 Loss = 0.959 Val loss = 1.189
Epoch = 391 Loss = 0.947 Val loss = 1.109
Epoch = 401 Loss = 0.930 Val loss = 1.101
Epoch = 411 Loss = 0.914 Val loss = 1.091
Epoch = 421 Loss = 0.958 Val loss = 1.091
Epoch = 431 Loss = 1.025 Val loss = 1.075
Epoch = 441 Loss = 0.916 Val loss = 1.083
Epoch = 451 Loss = 0.927 Val loss = 1.077
Epoch = 461 Loss = 0.888 Val loss = 1.067
Epoch = 471 Loss = 0.903 Val loss = 1.094
Epoch = 481 Loss = 0.918 Val loss = 1.080
Epoch = 491 Loss = 0.891 Val loss = 1.048
6. Results¶
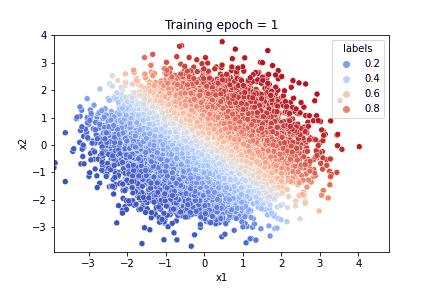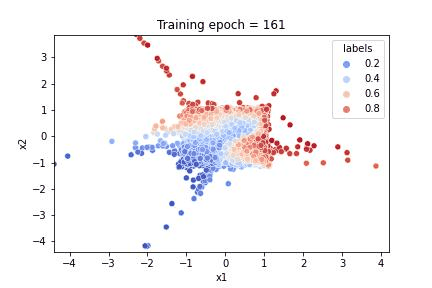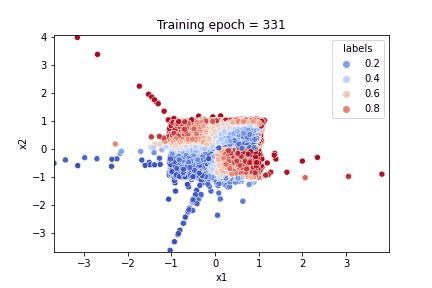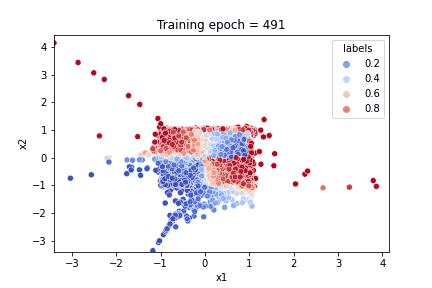
To illustrate the results of the training, we plot:
a gif illustrating how the algorithm learnt both to reproduce the feature distribution and the response
The loss and accuracy history
[13]:
from plot_utils import make_gif
make_gif(fig_list, "diglm_example.gif", duration=0.3)
[21]:
# loss plot
plt.figure(figsize=(13, 6))
plt.subplot(121)
plt.title('Training and validation loss')
plt.plot([i for i in range(len(history['train_loss'])-1)],
history['train_loss'][1:],
label='train loss')
plt.plot([i for i in range(len(history['val_loss'])-1)],
history['val_loss'][1:],
label='val loss')
plt.xlabel('Training steps')
plt.ylabel('loss')
plt.vlines([250], 1, 3.7, color='red')
plt.text(180, 3.5, r'$\lambda$ = 1. $\lambda$ = 0.5')
plt.grid(axis='y')
# accuracy plot
plt.subplot(122)
plt.title('Accuracy')
plt.plot([i for i in range(len(history['accuracy']))],
history['accuracy'])
plt.xlabel('Training steps')
plt.ylabel('accuracy')
plt.vlines([250], 0.35, 0.95, color='red')
plt.text(190, 0.4, r'$\lambda$ = 1. $\lambda$ = 0.5')
plt.grid(axis='y')
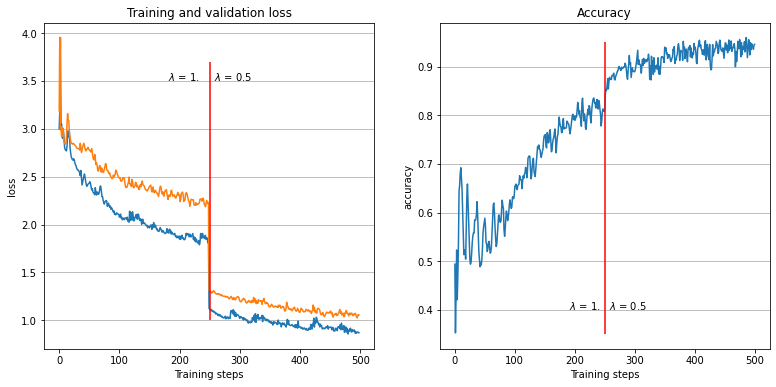
Referring to the gif, we see that the algorithm accomplished the task of evaluating the dataset feature distribution as well as the response function. The quality of the response evaluation is monitored through the BinaryAccuracy metric, counting the fraction:
where the Positive / Negative cut on the response function is set to 0.5. After 500 epochs, the loss function attains a plateau and the accuracy is stably around 0.94.
Just as in the SpQR notebook, we can evaluate the quality of the feature distribution evaluation using a Kolmogorov-Smirnov test
[22]:
from scipy.stats import ks_2samp
#slicing and preparing arrays to perform the test
mean, x1_trans, x2_trans = sampling(1000)
mean_target, x1_target, x2_target = np.squeeze(np.split(df.to_numpy(), 3, axis=1))
# calculating statistic from K-S test and p-values
stat_1, pval_1 = ks_2samp(x1_trans, x1_target)
stat_2, pval_2 = ks_2samp(x2_trans, x2_target)
print(f'p-value x1 distribution = {pval_1}')
print(f'p-value x2 distribution = {pval_2}')
p-value x1 distribution = 0.00026182909606191183
p-value x2 distribution = 0.00020538468989076796
We see that in this DIGLM model the capability of the algorithm to learn the univariate distributions of the features \(x_1\) and \(x_2\) is reduced with respect to the pure NeuralSplineFLow model (and we can expect it to be also less accurate in prediction than the simple GLM model). That’s because the algorithm tries to minimize the loss on a wider parameter space.
About \(\lambda\)¶
The hyper-parameter \(\lambda\) weights the two component of the log-likelihood loss, favouring the training of one part of the algorithm over the other. In middle of the training we switched it from 1 to 0.5: this resulted of course in a diminution of the likelihood, but what’s interesting to notice is that the binary accuracy atteined rapidly a better result after the diminution, just as stated in the article. To obtain pure predictive or pure generative model it is sufficient to set \(\lambda\) to very high (or very low) value (with respect to 1.).